Media
HKU biomedical engineering team develops a new bioinformatic tool for precise prediction of cells transformation and diseases formation including cancers
08 Feb 2022
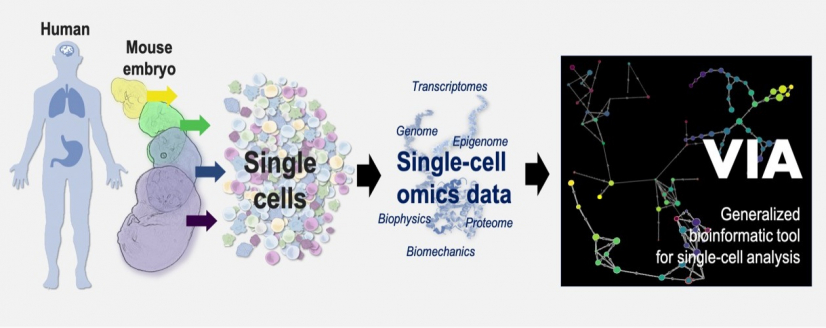
VIA is a generalised bioinformatic tool that helps the study on how the cells in human or animals change over time during normal development or diseases. VIA is widely adaptable to different important biological processes based on a wide range of data types including single-cell proteomic, transcriptomic, epigenomic, multi-omics and biophysical/ biomechanical datasets.
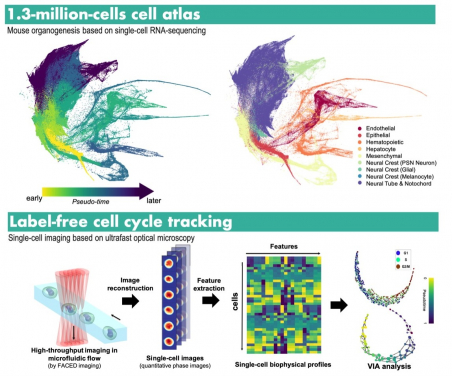
(Top) VIA can robustly uncover the intricate process at single cell precision that turns a single fertilised egg into a whole new individual with all the organs. (Bottom) Using the biophysical properties of single-cells extracted by an advanced microscope developed by the same team at HKU, VIA reveals subtle and variations of cell mass and the mass content distribution over the cell cycle progression - a fundamental cellular process that maintains life and might lead to cancer if the process goes wrong.
Cell is the basic unit of life. Over time, cells in human body constantly change to different forms. Even though it has been studied for more than a century, it remains challenging to understand what they do and how they change over time during normal health or ageing, or how these changes in cellular structure result in diseases including cancer.
Omics data analysis, which seeks to capture the rich information of single cells at high precision, is important for tracking cells transformation and hence for the prediction of occurrence and recurrence of diseases, and for survival analysis and discovery of biomarkers.
An interdisciplinary team led by Professor Kevin Tsia in the Department of Electrical and Electronic Engineering and Programme Director of Biomedical Engineering Programme at the University of Hong Kong (HKU) has developed a new computational tool for studying omics data that overcomes current challenges in the study of normal and malignant cell transformation.
Their latest breakthrough has recently been published in the academic journal Nature Communications.
“You can imagine cells are the mountain hikers traveling in a huge ‘landscape’. The fact that they evolve into different cell states and types in normal health and in disease can be analogous to that the hikers travel along different trails (or trajectories) in this landscape. Our ultimate goal is to find ways to locate cells on this landscape, predict their trajectories, and thus understand how our body develops normally and how it fails in diseases,” Professor Tsia explained.
“Accurate prediction of these trajectories relies on the ability to measure and analyse all the relevant signatures representing each cell. This is like performing in-depth interrogation with each hiker so that we can gather his/her identity, accessories, physical/mental conditions and so on,” he added.
A wide range of so called “omics technologies” are now available to capture rich information of single cells at high precision, such as a catalogue of genes that are “turned on/off” in cells or the protein types present in cells.
“However, current algorithms often cannot handle the sheer size of complex single cell omics data (more than millions of cells). They are also often designed to work well only for analysing only one specific type of cell signatures. Thus, we still lack a solution that is versatile enough to perform comprehensive trajectory inference,” noted Dr. Joshua Ho, Associate Professor in School of Biomedical Sciences in LKS Faculty of Medicine, HKU who also participated in this research work.
Overcoming these barriers, the new trajectory inference algorithm developed by the HKU team, called VIA, can process omics data up to 100 times faster than existing technologies, making it powerful in handling large-scale single-cell data (more than millions of cells). It can hence discover elusive cell lineages and rare cell fates in a variety of biological processes that can hardly be discovered by other methods, shedding important light into how disease evolves.
“Our method uses a novel unsupervised machine learning approach, which allows us to efficiently identify the best trajectories that represent the underlying biological progressions, no matter how complex the trajectories are. And we showed that VIA works very well, even when the cell population size is beyond what the current methods can handle,” explained by Shobana Stassen, who is the first author of the work from HKU Department of Electrical and Electronic Engineering and a recipient of the HKU’s Presidential PhD Fellowship.
To ensure that VIA can be widely adaptable, the team carried out comprehensive investigations of different important biological processes, based on a wide range of data types, including single-cell proteomic, transcriptomic epigenomic, as well as multi-omics datasets.
For examples, VIA can reveal complex and subtle transitions of cell state during haematopoiesis – a process of blood formation from bone marrow. It also robustly uncovered the fascinating and intricate process at a single cell precision that turns a single fertilised egg into a whole new individual with all the organs (a process called organogenesis), even when the cell count to be analyzed is beyond 1 million cells. This is the scale otherwise not affordable in many existing methods.
Another key outcome of this work is that the team explored the use of VIA to analyse the high-resolution cell image data that can infer physical traits of the cells - a cell signature that has largely been underexplored for trajectory inference.
Apart from the omics data, cell image contains rich information of how different cell types look differently and how the cell changes its outlook (morphology) over time.
Using an optical microscope system newly developed by Professor Tsia’s research lab, the team generated large amount of single cell image data at the speed at least 100 times faster than any typical microscopes. They then used VIA to successfully reveal subtle and variations of cell mass and the mass content distribution over the cell cycle progression - a fundamental cellular process that maintains life, and might lead to cancer if the process goes wrong. The development of this microscope was earlier published in Nature Protocols. (Aug 2021). A start-up company has been set up to transfer the technology for application of the new microscope system in bio-medical uses.
“VIA is an open-source tool and we made it publicly available. We are constantly improving and upgrading the tool. I hope VIA could widely benefit the community of biologists and biomedical scientists who investigate different aspects of biological evolution,” Ms Stassen added.
Meanwhile, some research teams in the US and the Mainland have adopted the VIA and the related computation tools developed by the team for COVID-19 research, to track and predict immune responses after infections or vaccinations, and the body’s response to treatments.
“Based on this groundwork, we are now applying VIA in different biomedical and biotechnology applications, in collaboration with researchers here in Hong Kong as well as overseas. Tip-of-the-iceberg examples include tracking anti-cancer treatment efficacy, as well as accelerating the drug discovery process in pharmaceutical industry, and many more,” said Professor Tsia, who was also awarded under the Research Grants Council’s Research Fellowship Scheme in 2020.
Please click here for more photos at the media conference and the presentation powerpoints.
The research paper “Generalized and scalable trajectory inference in single-cell omics data with VIA” was published in Nature Communications:
https://doi.org/10.1038/s41467-021-25773-3
The research paper published in Nature Protocol on the optical microscope system developed by Professor Tsia’s research lab:
https://www.nature.com/articles/s41596-021-00576-4 (Aug 2021)
Media enquiries:
Ms Celia Lee, Faculty of Engineering, HKU (Tel: 3917 8519; Email: leecelia@hku.hk) or
Mr Heng Cheng, Faculty of Engineering, HKU (Tel: 3917 1924; Email: hengc@hku.hk)