Media
HKUMed engineers highly accurate SaCas9 enzymes with single-base precision for therapeutic genome editing
09 Mar 2022
A research team of LKS Faculty of Medicine, The University of Hong Kong (HKUMed) has successfully engineered new Staphylococcus aureus Cas9 (SaCas9) variants, named KKH-SaCas9-SAV1 and SAV2, which, compared to the wild-type KKH-SaCas9, exhibit greatly enhanced targeting precision for editing only the disease-causing allele but not the normal allele. The new SaCas9 variants also reduce undesired off-target edits by more than 95% while possessing high editing activity. This study could open new possibilities to target disease-causing single-base mutations that occur in many genetic disorders. The findings are now published in Nucleic Acids Research [link to the publication] and a patent application has been filed based on this work.
Background
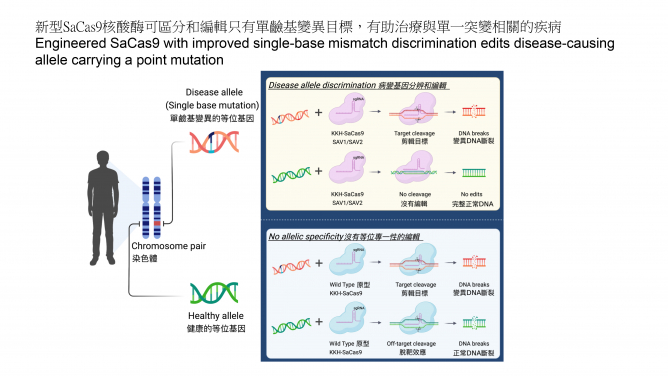
Advancing the genome editing tool makes more genetic diseases become treatable. CRISPR-Cas9 is a powerful genome editing tool by allowing targeted editing of DNA in living cells, and there has been a long-sought demand to apply this tool for tackling diseases with only single-base mutations. However, pre-existing Cas9 enzymes have limited capability in distinguishing normal and disease alleles that differ only by a single-base mutation in a patient. In this work, the HKUMed research team successfully expanded the applicability of Cas9 to target disease-causing single mutations (or single-nucleotide polymorphisms (SNPs)) and disable only the disease allele, while not affecting the healthy allele (Figure 1).
The ease of delivery, targeting range, efficiency and specificity are major considerations for choosing the right editing enzyme from the growing number of CRISPR systems for therapeutic genome editing. The team focused on engineering the KKH-SaCas9 enzyme, which is more compact than the commonly used Streptococcus pyogenes Cas9, and can be packed into adeno-associated viral vector for gene therapy applications. The KKH variant of SaCas9 chosen also has an expanded protospacer adjacent motif (PAM) compatibility than the wild-type SaCas9 for reaching more genetic loci.
Research findings
Though high-fidelity variants of the wild-type SaCas9 were developed for targeting of genomic sequences, the team found that previously reported mutations conferring higher specificity to the wild-type enzyme are not compatible with the KKH-variant, which has broader targeting range. Building on the previous work1 that showed how combining multiple mutations could optimise Cas9 enzymes for targeting accuracy and efficiency, the team searched for mutations in re-engineering and optimising the performance of KKH-SaCas9. To achieve this, the team used structure-guided combinatorial mutagenesis to screen through variants and identified two of them, KKH-SaCas9- SAV1 and SAV2, that outperform all other candidate variants in achieving greater genome-wide targeting specificity while exhibiting high editing activity in human cells. Specifically, KKH-SaCas9-SAV2 reduced off-target edits by more than 95%, while retaining over 80% of wild-type’s on-target editing activity. The research team will continue their engineering efforts to further boost the enzyme’s editing activity, as well as enable its use for mutation correction in addition to editing.
More importantly, the team revealed the unique capability of KKH-SaCas9- SAV1 and SAV2 in discriminating targets with single-base differences, including those locate distantly from the PAM site. This could expand the scope of genome editing to many single mutations and SNPs that could not have been targeted specifically. In therapeutic settings, this is critical to avoid targeting the highly similar normal allele. Using other SaCas9 variants, SNP-specific targeting confines only to those pathogenic SNPs or mutations that are located within the seed region of the guide RNA. This restrictive parameter has put limit on the range of genetic diseases that could be previously targeted.
Research significance
‘Our high-fidelity KKH-SaCas9 enzymes represent valuable new reagents that can be applied in precise editing of therapeutic targets for biomedical and in vivo genome editing applications. This could possibly allow genome editing to be applied in treating more genetic diseases in a safer way,’ said Dr Alan Wong Siu-lun, Assistant Professor of the School of Biomedical Sciences, HKUMed.
About the research team
This research was led by Dr Alan Wong Siu-lun, Assistant Professor of the School of Biomedical Sciences, HKUMed, as the corresponding author. Ms Chaya Yuen Tsz-lo, PhD student, Ms Dawn Thean Gek-lian, Research Assistant, Ms Becky Chan Ka-ching, PhD student, Dr Peng Zhou, Postdoctoral Fellow, Ms Cynthia Kwok Chui-shan, Research Assistant, School of Biomedical Sciences, HKUMed, were co-first authors, with assistance from Dr Chu Hoi-yee, Ms Maggie Cheung Sau-ha, Ms Wang Bei, PhD student, and Dr Gigi Choi Ching-gee, Postdoctoral Fellow. Other collaborators included Professor Anskar Leung Yu-hung of the Department of Medicine, HKUMed; Dr Zheng Zongli, Mr Chan Yee-man and Ms Silvia Mak from Ming Wai Lau Centre for Reparative Medicine, Karolinska Institutet, Hong Kong node.
Acknowledgement
This work was supported by the Centre for Oncology and Immunology under the Health@InnoHK Programme launched by the Innovation and Technology Commission, the Government of Hong Kong Special Administrative Region; Hong Kong Research Grants Council (TRS-T12-702/20-N); National Natural Science Foundation of China Excellent Young Scientists Fund (32022089). This work was also in part supported by the Ming Wai Lau Centre for Reparative Medicine Associate Member Programme.
Media enquiries
Please contact LKS Faculty of Medicine of The University of Hong Kong by email (medmedia@hku.hk).
1 Choi G.C.G., et al. Combinatorial mutagenesis en masse optimizes the genome editing activities of SpCas9. Nature Methods 16, 722-730 (2019)