Media
HKU Scientists Develop a Novel Approach to Interrogate Tissue-specific Protein-protein Interactions, Paving Ways to Understand Organ Development and Disease Pathogenesis
23 May 2024
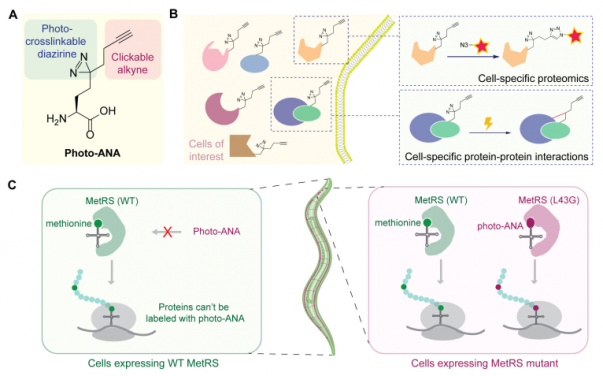
Cell-selective MACSPI analysis in C. elegans. (A) Chemical structure and functions of photo-ANA.
(B) A chemical proteomics approach to profile cell type-specific proteome and interactome. (C) Spatial selectivity is achieved by controlling the expression of the MetRS mutant, which can label proteins with photo-ANA, using cell-specific promoters in transgenic animals. Adapted from Huang et al., 2024, PNAS.
Multicellular organisms, like animals and plants, have complex cells with diverse functions. This complexity arises from the need for cells to produce distinct proteins that interact with each other. This interaction is crucial for cells to carry out their specific tasks and to form complex molecular machinery. However, our current understanding of such protein-protein interactions often lacks cellular contexts because they were usually studied in an in vitro system or in cells isolated from their tissue environment. Effective methods to investigate protein-protein interactions in a tissue-specific manner are largely missing.
To bridge this technology gap, a collaborative research team of The University of Hong Kong (HKU), led by Professor Xiang David LI from the Department of Chemistry and Professor Chaogu ZHENG from the School of Biological Sciences, both from the Faculty of Science, along with Dr Xiucong BAO from the School of Biomedical Sciences, Li Ka Shing Faculty of Medicine, recently developed a novel chemical biology approach to label proteins from specific cells with a bifunctional amino acid probe that allows labelled proteins to be isolated and captures protein-protein interaction through photo-crosslinking. This new method, Methionine Analog-based Cell-Specific Proteomics and Interactomics (MACSPI), uses a chemical probe to label proteins in specific cells and capture their interactions. Using this approach, the team has identified many new tissue-specific proteins and protein interactions, helping us better understand how cells work in living organisms and study various biological problems, such as organ development and disease pathogenesis. The research work was recently published in a leading multidisciplinary journal – Proceedings of the National Academy of Sciences (PNAS).
Innovative design
The team designed and synthesised an unnatural amino acid (photo-ANA) that is structurally similar to methionine, the naturally occurring amino acid, but with two additional components (see Figure 1). One component is an alkyne group, which can be used as a chemical handle for the labelled proteins to be extracted and purified. The other is a diazirine group, which can be activated by light to create stable covalent linkages between the labeled proteins and any molecules they interact with. Next, the team engineered an enzyme called MetRS to create a variant that can recognise and incorporate the unnatural amino acidinto proteins as they are being built. By controlling the expression of this engineered enzyme in specific tissues, only proteins from the tissue of interest are labelled by chemical probe. Moreover, with light-induced crosslinking, protein complexes from specific tissues can be captured and isolated.
As a proof-of-concept, the team applied the MACSPI method to profile proteins from muscle cells and neurons, respectively, in a model organism called C. elegans and found many novel tissue-specific proteins. The team also demonstrated the method’s utility in capturing tissue-specific protein-protein interaction by identifying tissue-specific interactors of a ubiquitously expressed protein, such as the molecular chaperone called HSP90. It was found that HSP90 binds to distinct sets of proteins to regulate different biological processes in muscles and neurons.
"This study is an excellent example of how innovative chemical labelling methods can help solve difficult biological problems," said Professor Xiang David Li.
"Understanding protein-protein interaction at the cellular resolution is often critical to decipher the molecular mechanism of a pathological process. For example, we are currently exploring the functions of the neuronal HSP90 interactors we identified; some appear to be involved in neurodegeneration in a Parkinson's disease model," said Professor Chaogu Zheng. The team envisions that the MACSPI method can be used in many multicellular organisms to profile proteomes and interactomes with spatial and temporal specificity, which can facilitate a broad spectrum of biological and biomedical research.
About the research team
This study is a collaboration among Professor Xiang David Li's team at the Department of Chemistry (Faculty of Science), Professor Chaogu Zheng's team at the School of Biological Sciences (Faculty of Science), and Dr Xiucong Bao's team at the School of Biomedical Sciences (Li Ka Shing Faculty of Medicine), The University of Hong Kong. Postdoctoral research fellow Dr Siyue Huang from Professor Li's team and research postgraduate student Ms Qiao Ran from Professor Zheng's team are co-first authors of the study. This work was initially supported by seed funds from the Strategic Interdisciplinary Research Scheme at HKU and subsequently supported by grants from the Hong Kong Research Grants Council Collaborative Research Fund, the Areas of Excellence Scheme, and the General Research Fund, Food and Health Bureau, and the National Natural Science Foundation of China.
About the research paper: “Huang, S.#, Ran, Q.#, Li, X.-M., Bao, X.*, Zheng, C.*, and Li, X.D.* MACSPI Enables Tissue-Selective Proteomic and Interactomic Analysis in Multicellular Organisms. Proc. Natl. Acad. Sci. U.S.A. 2024 May 13”
The journal paper can be accessed from here: https://www.pnas.org/doi/10.1073/pnas.2319060121
For media enquiries, please contact Ms Casey To, External Relations Officer (tel: 3917 4948; email: caseyto@hku.hk) / Ms Cindy Chan, Assistant Director of Communications of HKU Faculty of Science (tel: 39175286; email: cindycst@hku.hk).
Images download and captions: https://www.scifac.hku.hk/press
