Media
Spy in protein ‘social networks’— HKU Chemists develop new chemical tools to study protein-protein interactions in living cells
07 Jan 2016
 Dr Li Xiang’s research team and collaborator at HKU Department of Chemistry: (from the left) Bao Xiucong, Li Xiaomeng, Dr David Li Xiang, Yang Tangpo and Dr Fung Yiman Eva.
Dr Li Xiang’s research team and collaborator at HKU Department of Chemistry: (from the left) Bao Xiucong, Li Xiaomeng, Dr David Li Xiang, Yang Tangpo and Dr Fung Yiman Eva.A research group led by Dr David Li Xiang from Department of Chemistry, the University of Hong Kong, has developed a novel chemical tool for deciphering complex protein networks of a cell — the elementary unit of life. The disorder of such cellular networks often leads to severe human diseases such as cancer and Alzheimer's diseases. The findings were recently published in a top-class scientific journal — Nature Chemical Biology. This is the first time that a study led by Hong Kong researchers is published in this prestigious journal.
The main components of a cell are macromolecules called proteins; they are chief actors of most cellular processes and determine the structure and function of the cell. Like most jobs in human society, the ‘jobs’ in the cell cannot be done by one single protein, but require a teamwork of thousands of different proteins, which physically interact to form protein-protein interaction networks or protein ‘social networks’. Errors in the regulation of these protein interactions can cause severe human diseases such as cancer. The mapping of proteins and their interactions are thus a fundamental challenge in modern biology with important applications in disease diagnosis and therapy.
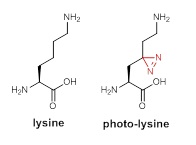
Unfortunately, proteins do not use Facebook or Twitter, making it difficult to know proteins’ ‘social networks’. For many years, researchers have been looking for a reliable method to discover cellular protein-protein interactions. To tackle this challenge, Dr Li’s group decided to send a ‘spy’ into cells for surveillance of cellular protein interactions. They designed and synthesized an unnatural amino acid called photo-lysine. Because of its high similarity with lysine, one of the 20 natural amino acids — the building blocks of proteins, photo-lysine serves as an excellent ‘undercover agent’ that easily deceives cellular protein synthesis machinery, so as to be incorporated into proteins without disturbing normal cell function. Importantly, photo-lysine also carries a light-activated ‘spy camera’ — diazirine group, which is able to ‘capture’ every interacting partner of a protein of interest in the cell when being exposed to ultraviolet (UV) light.
As a research tool, photo-lysine is also very easy to use. In the published paper, Dr Li’s group has demonstrated that by simply adding photo-lysine into the cell culture medium for around 16 hours, the cellular proteins of the human cells were sufficiently labeled by this ‘spy’ unnatural amino acid. They then exposed the photo-lysine-labeled cells to UV light for a couple of minutes to ‘fix’ or ‘trap’ all the cellular protein-protein interactions. When coupled with state-of-the-art mass spectrometry-based proteomics approaches, Dr Li’s lab successfully identified many known and undiscovered protein interactions that are important for the regulation of essential cellular processes such as gene expression, signal transduction and metabolism. With this new chemical tool, scientists can now map mysterious proteins interaction networks in cells, which will help us to gain more comprehensive understanding of human disease and provide direction for therapy development.
About the paper
“Photo-lysine captures proteins that bind lysine post-translational modifications”
at Nature Chemical Biology
http://www.nature.com/nchembio/journal/vaop/ncurrent/full/nchembio.1990.html
Nature Chemical Biology is a top-class international scientific journal publishing original research from the expanding community of chemical biologists.
About Dr David Li Xiang
More information about Dr Li Xiang David and his research group can be found from their group’s webpage: http://www.chemistry.hku.hk/staff/xiangli/xiangli/index.html
For press enquiry, please contact Ms Cindy Chan, Senior Communication Manager of Faculty of Science, at 3917-5286/ 6703- 0212 or by email at cindycst@hku.hk
Image download
http://www.scifac.hku.hk/news/media?page=1